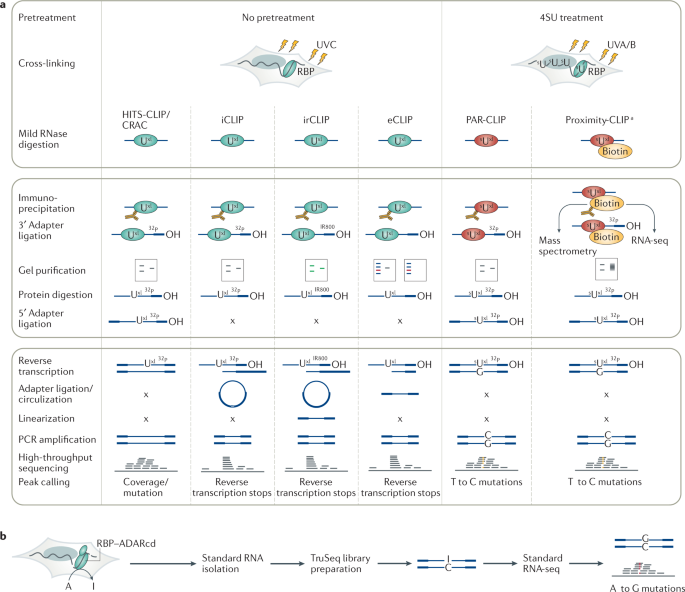
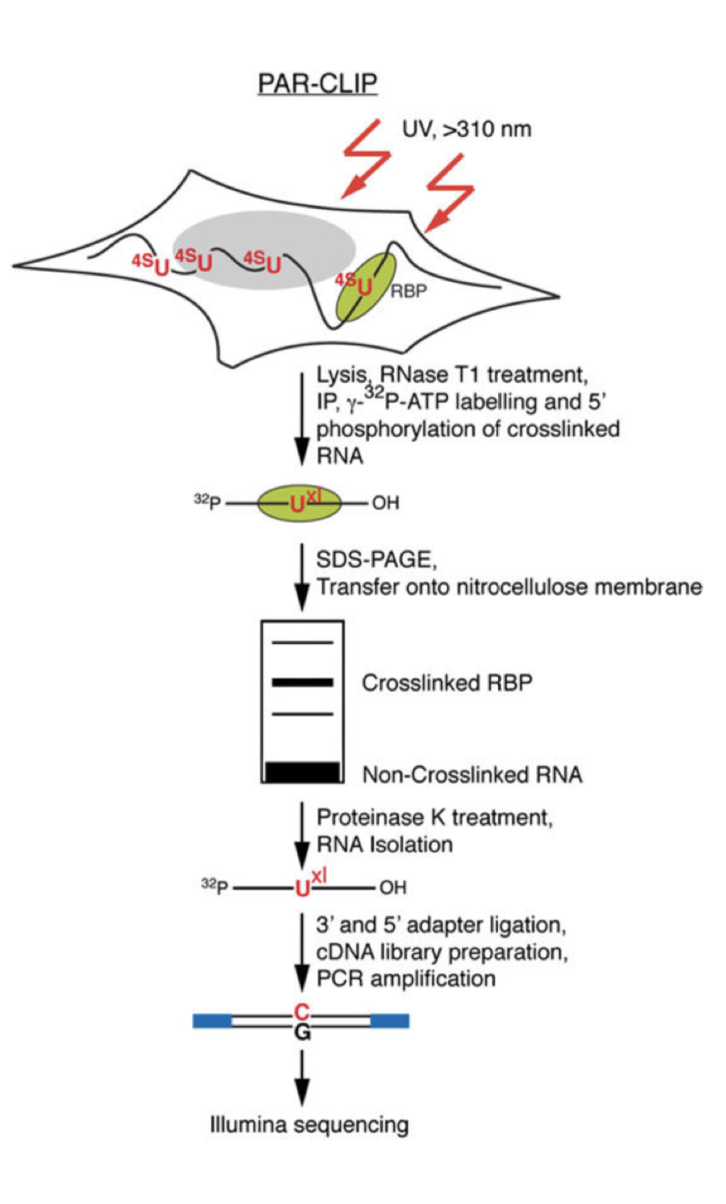
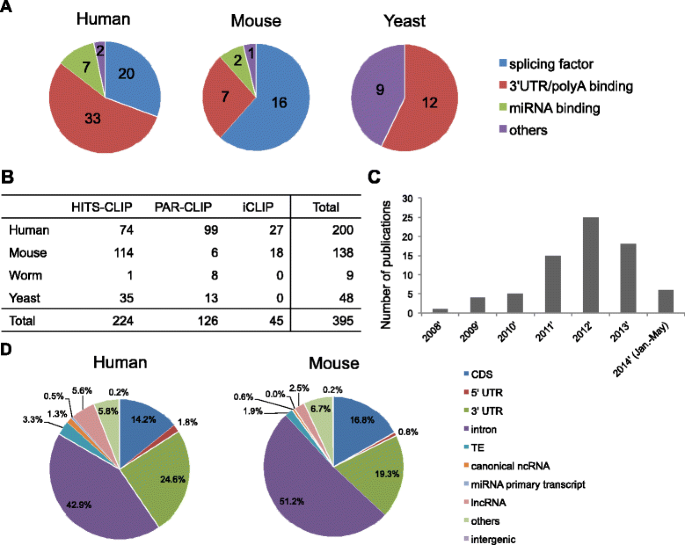
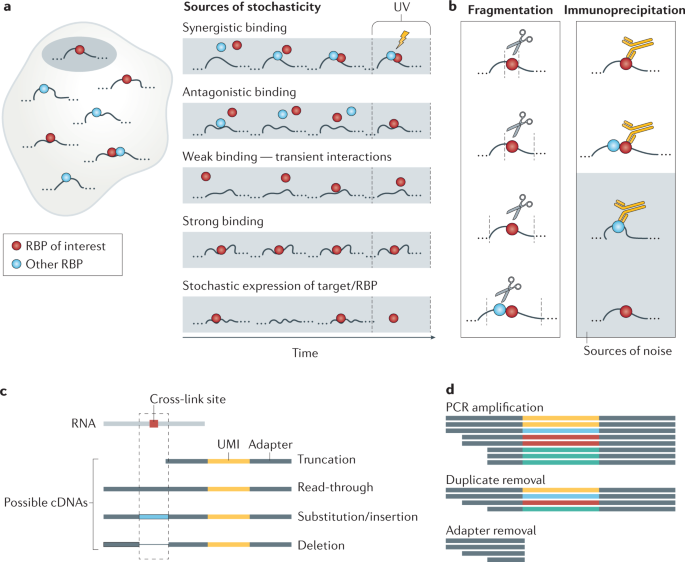
Optimization of PAR-CLIP for transcriptome-wide identification of binding sites of RNA-binding proteins - ScienceDirect
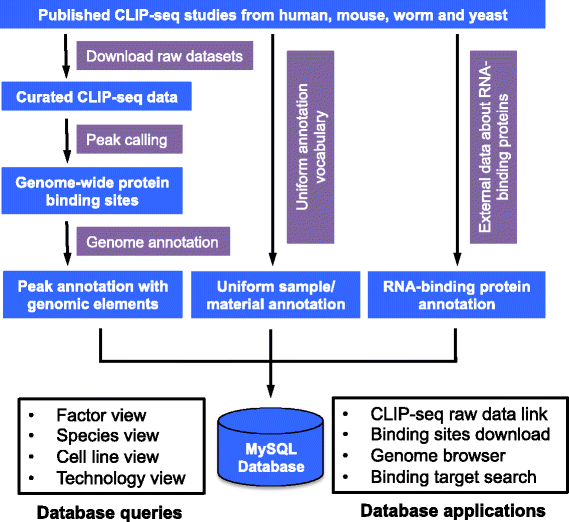
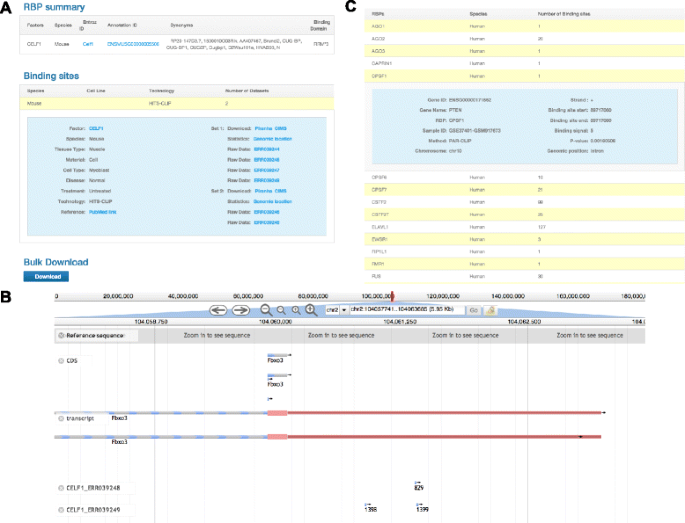
PRAS: Predicting functional targets of RNA binding proteins based on CLIP-seq peaks | PLOS Computational Biology
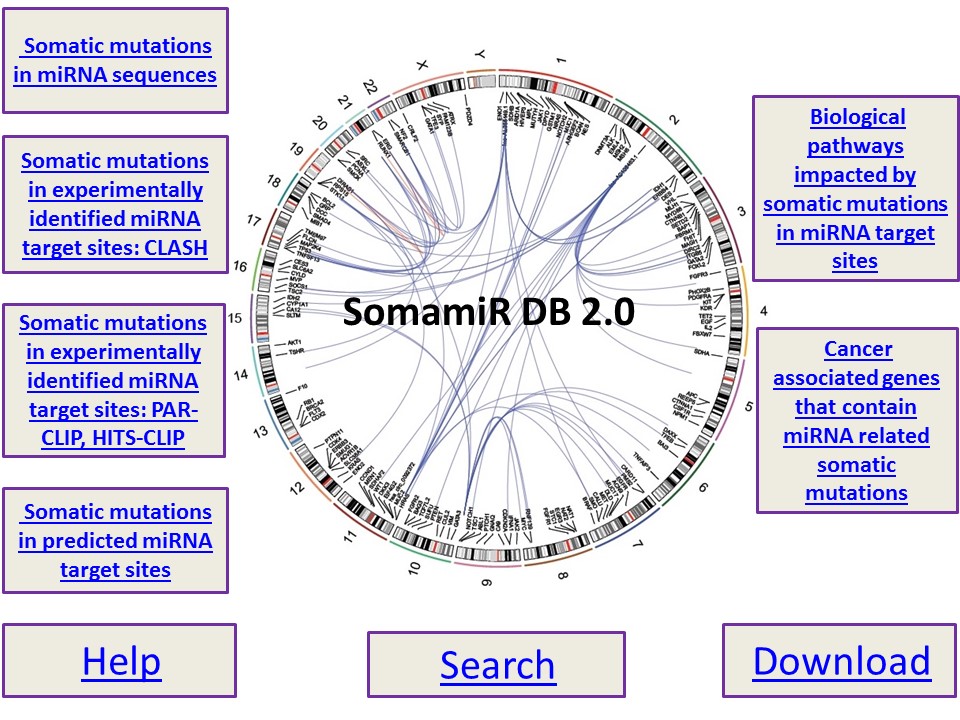
![PDF] PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways | Semantic Scholar PDF] PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/f09afb70d8127227f77a4ec068d17ec7563493c8/3-Table1-1.png)
PDF] PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways | Semantic Scholar
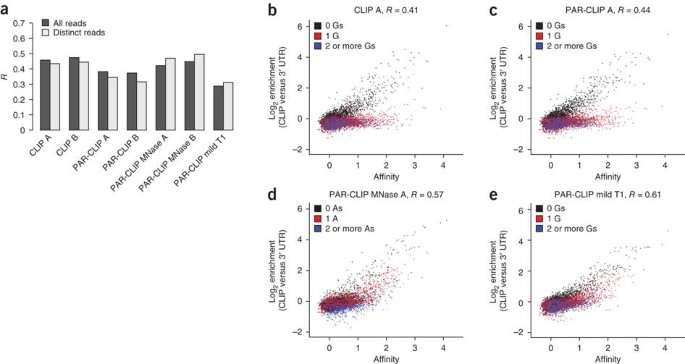
A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins | Nature Methods

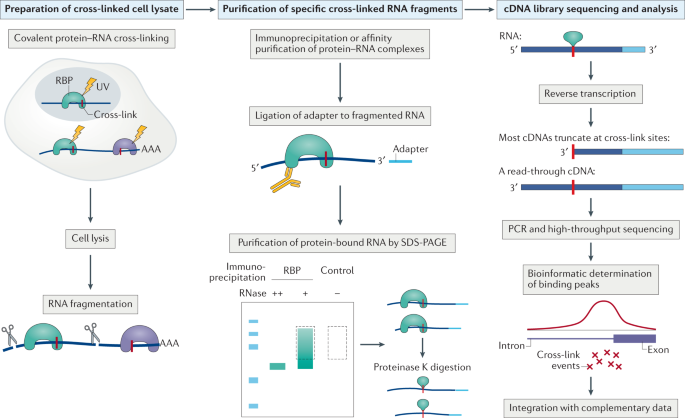
PAR-CLIP: A Method for Transcriptome-Wide Identification of RNA Binding Protein Interaction Sites | SpringerLink

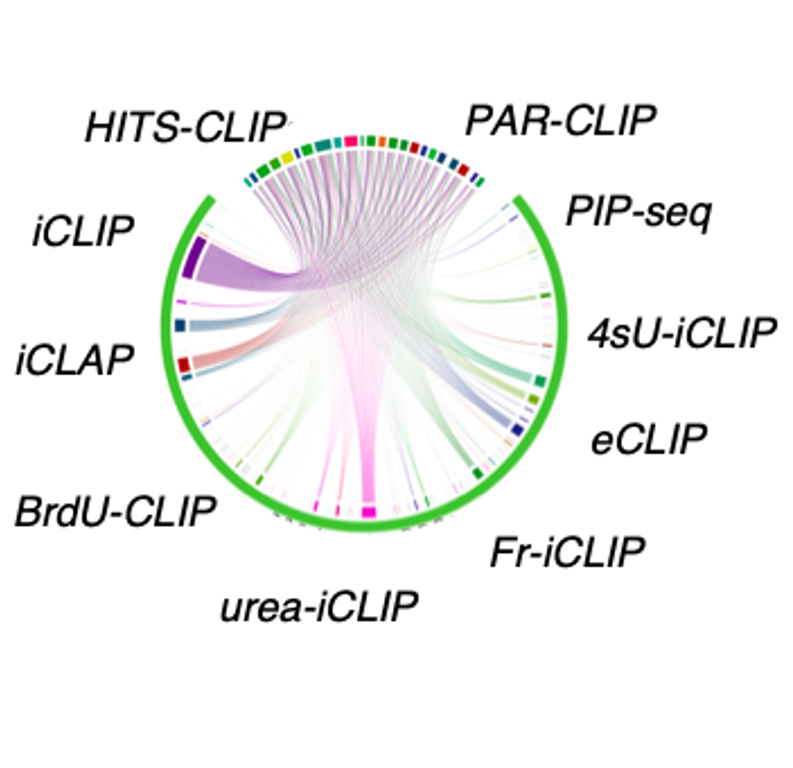
Outline of HITS-CLIP, PAR-CLIP and several variants, iCLIP, iCLAP and... | Download Scientific Diagram